Open Science Repository Agriculture
doi: 10.7392/Agriculture.70081945
An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers
M. A., Farah Fazwa [1], H., Siti Salwana [1], H., Maideen [2], O., Mohamad [3]
[1] Forestry Biotechnology Division, Forest Research Institute Malaysia, 52109 Kepong, Selangor, Malaysia
[2] Faculty of Science and Technology, University Kebangsaan Malaysia, 43600, Bangi, Selangor, Malaysia
[3] Faculty of Plantation and Agrotechnology, Mara Technology University, (UiTM), 40450, Shah Alam, Selangor
Abstract
Labisia pumila, locally known as Kacip Fatimah, and member of the Myrsinaceae family, is one of the popular herbal species in Malaysia. The leaf or the whole plant is traditionally used to treat internal problems and women’s health. The objective of this study was to evaluate the genetic relationships among superior accessions for two varieties of L. pumila (L. pumila var. pumila and L. pumila var. alata) using amplified fragment length polymorphism (AFLP) technique. Superior accessions of both varieties were selected based on their total phenolic contents, ranging from 2597 to 2928 mg/50 g GAE (gallic acid equivalent), which had been determined during a previous work. Three primer combinations that produced clear scorable and highly polymorphic bands were screened from 64 EcoRI and MseI primer combinations. The numbers of AFLP fragments generated per primer set ranged from 30 to 124 with fragment sizes varying from 50 to 500 bp. A total of 170 AFLP fragments were detected and all are polymorphic. AFLP banding patterns were then transformed into presence–absence binary data and matrices were processed with NTSys software programs. Results showed that the genetic relatedness for L. pumila var. pumila and L. pumila var. alata was not close at J=0.300 and J=0.298, respectively. No close genetic relatedness was also recorded between the two varieties at J= 0.260. Dendogram based on UPGMA method demonstrated that the two varieties fell into different clusters. The important finding of this study is the identification of genetic relationships among the 119 accessions of both varieties with high total phenolic content which can be used for future breeding activities.
Keywords: Labisia pumila var. pumila, L. pumila var. alata, total phenolic content.
Citation: Md Ariff, F. F., Hashim, S. S., Haja, M., & Osman, M. (2013). An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers. Open Science Repository Agriculture, Online(open-access), e70081945. doi:10.7392/Agriculture.70081945
Received: February 25, 2013.
Published: March 22, 2013.
Copyright: © 2013 Md Ariff, F. F., Hashim, S. S., Haja, M., & Osman, M. Creative Commons Attribution 3.0 Unported License.
Contact: [email protected]
Introduction
Labisia pumila, commonly known as Kacip Fatimah from the family Myrsinaceae, is one of the popular herbal species in Malaysia and well recognized by Malay women for its medicinal value. The leaf or the whole plant is traditionally used to treat internal problems and women’s health, especially to induce and facilitate childbirth as well as a post-partum medicine (Burkill 1993). The other uses of this herb are treatment for dysentery, dysmenorrhea, flatulence, and gonorrhea (Muhamad & Mustafa 1994). In addition, it is also used for rheumatism, indigestion, reducing menstrual pain in treatment of hemorrhoids and as female aphrodisiac (Fasihuddin & Hasmah 1993). It is a popular herb that has long been recognized to contain high bioactive compounds (Jamal Azdina et al. 1998, Jaafar et al. 2008) and is demanded for its medicinal value as female tonic, health and herbal products.
Even though the plant is highly demanded, most of the raw materials for the development of herbal products in the market are being collected from the wild, without knowing the quality of the materials. This condition, therefore, directly affects the quality of the products. Hence, research on the screening of superior accessions of two varieties of L. pumila based on their high total phenolic contents has been carried out (Farah Fazwa et al. 2010, 2012). One hundred and fifty accessions (for each variety) were collected wild from ten natural populations in Peninsular Malaysia and screened for their total phenolics using the Folin-Ciocalteu method (Rossi & Singleton 1965). Recently, it was found that the bioactive compounds of L. pumila consist of resorcinols, flavonoids and phenolic acid (Jamia Azdina 2004, Jaafar et al. 2008). These compounds contribut to the antioxidative properties that may reduce oxidative damage to the human body (Scalbert & Williamson 2000). Natural antioxidants constitute a broad range of compounds including phenolic, nitrogen and carotenoid compounds (Velioglu et al. 1998). Plant antioxidants are believed to play a role in protection against a variety of diseases and to delay ageing processes (Wong et al. 2006). As a result, a total of 62 accessions have been identified as superior which are able to produce high total phenolic contents that range from 2597 to 2928 mg/50 g GAE (gallic acid equivalent). Both L. pumila varieties contain phenolic contents of more than 1000 mg/50 g, thereby qualifying them as plants with high phenolic contents as classified by Vimala (2010) (pers. comm). Determination of genetic variation by RAPD method based on total phenolic content for four forms of Impatiens balsamina has also been conducted by Nurul et al. (2010). These superior plants will be used for selection work in future as part of a breeding strategy for the species. Even though many studies have been done on the screening of total phenolics from this species (Ehsan et al. 2011, Lee et al. 2011, Mohd Hafiz & Hawa 2011a, 2011b), the genetic relationships among the superior accessions using genetic marker have never been reported. Through literature search, it is found that only a few studies have been done on the identification of L. pumila using molecular markers. Isozyme markers have been used to study the genetic diversity of L. pumila populations in humid tropical forests (Wickneswari et al. 2000) and DNA-based analysis like random amplified polymorphic DNA (RAPD) have been successfully used for the characterization of three varieties of L. pumila from nine accessions in two states of Peninsular Malaysia by Bhore et al. (2009). However, amplified fragment length polymorphism (AFLP) (Vos et al. 1995), one of the DNA marker techniques, has never been tested before to this species. AFLP technique is an effective, cost-efficient and reproducible method for revealing DNA polymorphism without any prior knowledge of the genome of the species being studied (Mueller & Wolfenbarger 1999). It is a reliable method of genetic fingerprinting and has been successfully used for characterization and evaluation of genetic relationship in many crops (Chen et al. 2004). Therefore, in the present study AFLP analysis was used to evaluate the genetic relationships among 62 superior accessions of two varieties of L. pumila based on genetic similarity.
Materials and method
Plant materials & DNA extraction
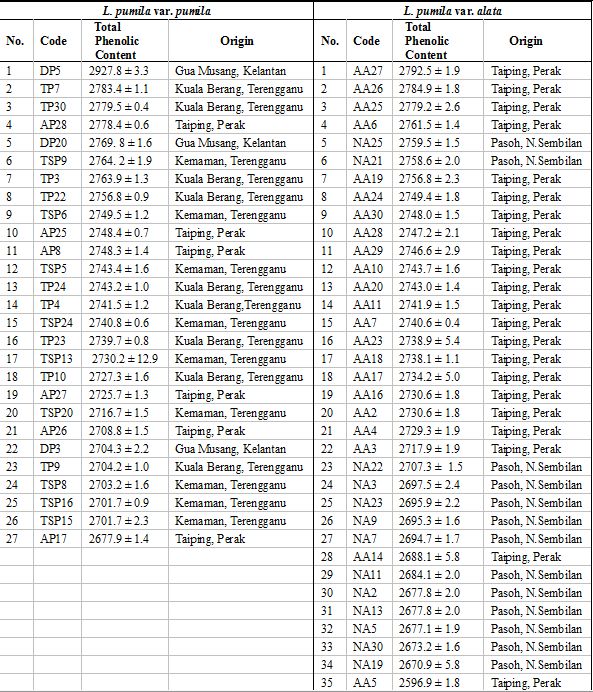
A collection was made of total of 62 superior accessions from both varieties which originated from six natural populations located in four states of Peninsular Malaysia, namely four populations of L. pumila var. pumila from i) Bukit Larut Forest Reserve (FR), Taiping, Perak, ii) Tembat FR, Kuala Berang Forest Reserve, Terengganu iii) Sungai Nipah FR, Kemaman, Terengganu and iv) Batu Papan FR, Gua Musang, Kelantan for L. pumila var. pumila; and two populations of L. pumila var. alata from i) Pondok Tanjung FR, Taiping, Perak, and ii) Pasoh FR, Jelebu, Negeri Sembilan (Table 1). All accessions were tagged with different code numbers and are being maintained in a germplasm bank at the Forest Research Institute Malaysia (FRIM), Kepong, Selangor, Malaysia. A total of 62 (27 L. pumila var. pumila and 35 L. pumila var. alata) superior accessions with their total phenolic contents are listed in Table 2. Young leaves from each superior genotype were collected for DNA extraction. Genomic DNA was extracted using the modified Murray and Thompson (1980) method and further purified using High Pure PCR Template Preparation Kit (Roche Diagnostics).
Table 2: List of 62 superior accessions for the two varieties of Labisia pumila used in this study.
(click to get access to larger version)
Note: Total phenolic content were expressed in unit of mg/50 g gallic acid equivalent (GAE) (Source: Farah Fazwa et al. 2012).
Amplified fragment length polymorphism (AFLP) analysis
About 250 ng of genomic DNA was double-digested with 2.5 U of EcoRI and MseI for 2 h at 37 °C. AFLP analyses were performed following the manufacturer’s instructions from the AFLP Reagent Kit (Applied Biosystem). The resulting template fragments were ligated to the adapters specific for the EcoRI and MseI restriction sites and a pre-amplification reaction was then carried out with EcoRI+A and MseI+C primers. The second PCR (selective amplification) was performed with primer pairs having three additional selective nucleotides at the 3′ end. With fluorescently labelled EcoRI+3 primers the PCR products were separated using 3131 Genetic Analyser. Out of 32 combine primers, three primers were selected which produced reliable amplification patterns.
Data analysis
Amplified DNA fragments were automatically scored with Gene Mapper 4.0 software (Applied Biosystem) and the binary data matrices were constructed. These data matrices were analysed using NTSYS-pc software (Numerical Taxonomy and Multivariate System) (Rohlf 1993). Percentages of polymorphic bands and polymorphic information contents were calculated. Genetic similarity among all selected accessions was calculated based on the AFLP data using Jaccard’s similarity index (Jaccard 1908) by the SIMQUAL subprogram. The SAHN subprogram was used for cluster analysis by the UPGMA method (unweighted pair-group method with arithmetic means) (Sneath & Sokal 1973).
Results and discussion
Polymorphic analysis of the primer combinations
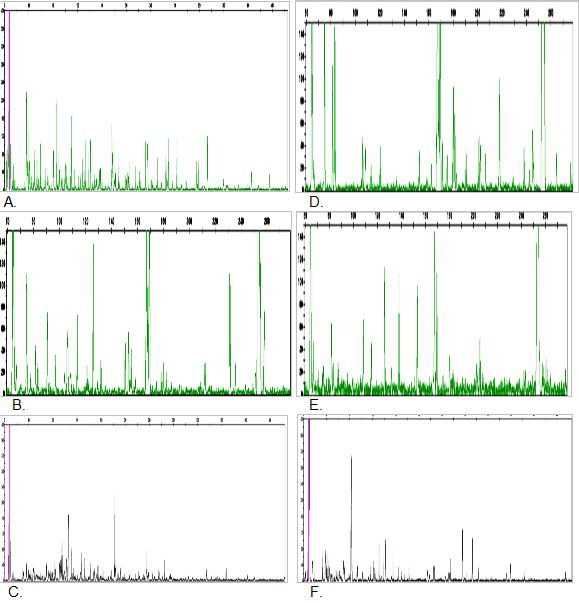
In this study, a total of170 scorable fragments ranging in size from 50 to 500 bp were detected using three primer combinations (Table 3). The average number of bands per primer combination was 56.6, of which 100% were polymorphic. The largest number of polymorphic bands (124) was produced with primer combination E-AGC/M-CAC, and the least number of polymorphic bands (16) was detected using primer combination E-ACG/M-CAG. PIC values ranged from 0.812 to 0.888 (mean 0.892). Some representative AFLP patterns of the two varieties are given in Figure 1. The results are comparable with those of a previous study by Bhore et al. (2009), which produced a total of 72 bands ranging 250 - 3000 bp assayed with RAPD marker technique using two primers (OPA1 and OPA2) for nine accessions of L. pumila. From the amount, 59 bands were polymorphic (82%) and 13 bands were not polymorphic (18%). The findings explained the use of AFLP markers as a powerful tool by revealing higher numbers of maximum bands and polymorphic bands when compared to RAPD technique. Other than that, the sample sizes used in these two different studies could also be a factor for these differences (Kularatne 2000). The number of samples analysed in the previous study was limited as compared with this study, which involved a high number of samples and covering seven populations throughout Peninsular Malaysia. The study by Gowda et al. (2010) found a total of 162 maximum scorable fragments from six primer combinations for authentication of the genus Embelia (Myrsinaceae) which comes from the same family as L. pumila using AFLP markers. These results have similar trend to the present study in terms of number of scored DNA fragments. They are also comparable with the findings for other herbal species such as Curcuma sp. which was reported to produce 202 amplified fragments, of which, 158 bands (78.2%) were polymorphic generated from nine AFLP primer combinations (Keeratinijakal et al. 2010).
Table 3: AFLP primer combinations, total numbers of fragments generated by each primer set, numbers of polymorphic fragments, percentages of polymorphic fragments and polymorphic information content (PIC) values used in the study of 62 superior accessions of L. pumila.
Figure 1: AFLP patterns of L. pumila var. pumila (A, B & C) and L. pumila var. alata (D, E & F) using primer pairs EACG/MCAC, EACG/MCAG and EAGC/MCAC. X-axis = base pair of alleles; Y-axis = intensity of alleles.
(click to get access to larger version)
Genetic similarity within and between varieties of all superior accessions
Traditionally, plant breeders have always evaluate plant origins and pedigrees as an extra indication of the characteristics that are acquired by a new genotype. With the advent of molecular techniques, new tools that have the capacity to reveal similarities in the genome of related plant species or cultivars directly become available. Knowledge of genetic similarity between accessions is useful in any breeding programme because it facilitates efficient sampling and utilization of germplasm resources. The breeder can use genetic similarity information to make informed decisions regarding the choice of accessions to cross for the development of populations or to facilitate the identification of diverse parents to cross in hybrid combinations in order to maximize the expressions of heterosis (Ajmone et al. 1998).
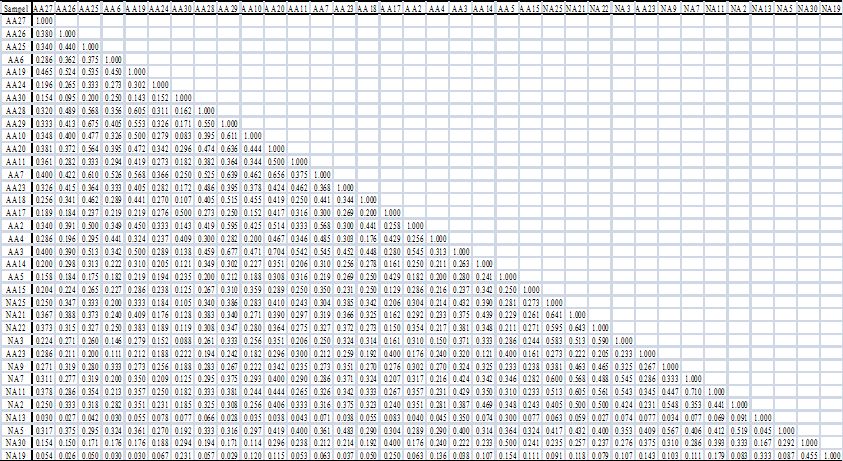
Genetic similarity coefficients between pairs of accessions based on Jaccards’s coefficient are presented in Table 4 and Table 5. For L. pumila var. pumila, the greatest genetic similarity (0.647) was observed between accessions TSP20 and TSP13, while the lowest genetic similarity (0.027) was observed between accessions TSP15 and TP10, with mean similarity of 0.300. For L. pumila var. alata the greatest genetic similarity (0.704) was observed between accessions AA3 and AA20, while the lowest genetic similarity (0.026) was observed between accessions NA19 and AA26, with mean similarity was 0.026. The mean similarity between both varieties was 0.260. The results indicate that all the selected accessions are genetically not close within and between varieties.
The difference in genetic similarity within the variety was probably due to the sample collection done within the same population. During the collection, the distance between one accession and another was far apart. Therefore, the possibility to acquire similar genetic constituents within the same population was low. In addition, the pollinators involved in pollinating herbal plants including L. pumila are those of a shorter distance where the pollination coverage area is limited (Michael et al. 2000). This explains that all accessions in the same population might not necessarily have similar genetic constituents and therefore the value of genetic similarity was recorded low. The genetic similarity was also low (0.26) between the two varieties. The value confirms that both varieties were far apart from each other. Besides extensive morphological differences between both varieties, the similarity indices also indicate that both varieties are distinct. From the results, it is suggested that AFLP markers may be used as dependable identifying parameters for varieties identification even if samples are collected from different locations. The results also explain that genetic differentiation is very much affected by geographical distances (Botanga et al. 2002, Yang et al. 2007).
Table 4: Genetic similarity indices for L. pumila var. pumila superior accessions.
(click to get access to larger version)
Table 5: Genetic similarity indices for L. pumila var. alata superior accessions.
(click to get access to larger version)
Cluster analysis and genetic relationship among accessions
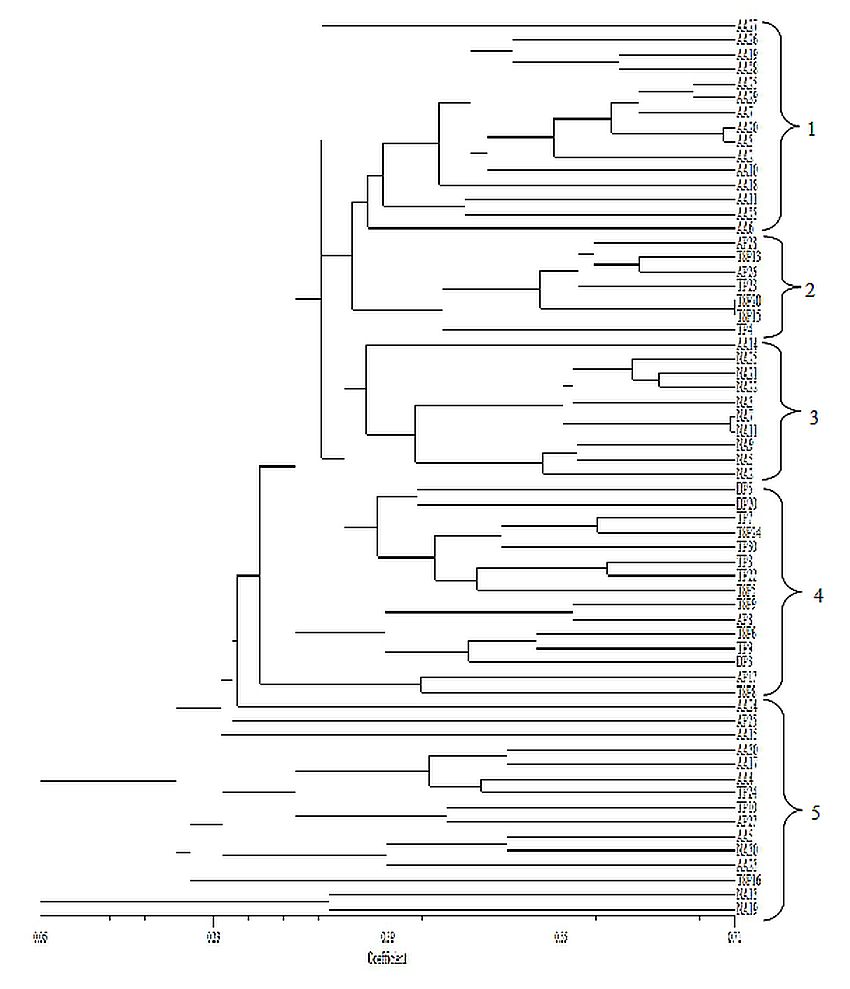
The dendogram produced by the UPGMA cluster analysis of the pooled AFLP data from the three combination primers is presented in Figure 2. The dendogram from the analysis demonstrated that L. pumila var. pumila and L. pumila var. alata were grouped into five main clusters, which is in agreement with their morphological traits. The first and third clusters only included L. pumila var. alata. However, for the first cluster, it grouped only 15 accessions from Pondok Tanjung FR, Taiping, whereas for the third cluster most of the accessions (9 out of 10) were from Pasoh FR, Jelebu. The second and fourth clusters only included L. pumila var. pumila. In the second cluster, seven L. pumila var. pumila accessions from Bukit Larut FR, Taiping; Sungai Nipah FR, Kemaman; and Tembat FR, Kuala Berang; were grouped together. Cluster four formed majority (13 out of 15) a group of accessions that originated from the eastern area of Peninsular Malaysia which is from the states of Kelantan and Terengganu. The final cluster comprising both of L. pumila varieties, were five accessions from L. pumila var. pumila and 10 accessions from L. pumila var. alata. In the results of a previous work on the dendogram produced using RAPD markers, three varieties of L. pumila, namely L. pumila var. pumila, L. pumila var. alata dan L. pumila var. lanceolata, were divided into two major clusters (Bhore et al. 2009). However in the study, the number of samples used is very limited which involved only nine accessions of L. pumila collected randomly from Melaka and Negeri Sembilan states of Malaysia.
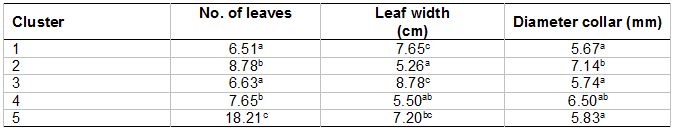
To the best of our knowledge, this is the first report on the application of the AFLP technique in the evaluation of the genetic relationships in L. pumila varieties. Even though the results indicated that all the superior accessions come from the same variety did not completely belong to one cluster, however the dendogram constructed on the basis of genetic similarity showed that all the L. pumila accessions clustered in a way that is generally in agreement with the classification based on their morphological characters. The difference among the two varieties had been further clarified by the assessment of some morphological characteristics as shown in Table 6. Morphological data clearly showed that there are significant differences between the two varieties. However, the cluster with mixed varieties (Cluster 5) is not significantly different from the other four clusters especially in the leaf width and diameter collar. This is because the cluster comprised a mixture of both varieties. These materials will be of great value to future genetic gains in cultivar productivity.
Figure 2: Dendogram of 62 high yielding accessions of Labisia pumila varieties. The designations refer to accessions listed in Table 2.
(click to get access to larger version)
Table 6: Morphological data for the five clusters.
Note: Means followed by the same letters within the same column are not significantly different at p ≤ 0.05.
Based on the findings, it is suggested that the identification of varieties should be further supported by their morphological and physiological data. Furthermore, L. pumila plants have high similarity in morphological characteristics, especially the shapes of their leaves and petioles, which may cause confusion in their utilization. However, identification based on morphological and physiological traits requires a large set of phenotypic data that are often difficult to obtain and sometimes vary due to environmental influences, which may lead to unreliable or erroneous determination (Barracosa et al. 2008).
Conclusion
Clear separation of all 62 accessions proved that an AFLP technique is a useful tool for future identification in L. pumila varieties. The low genetic similarity within and between varieties showed that each of the individuals is separately and genetically not close related to each other. Whenever there is genetic distance, it indicates that genetic constituents between plants are different. The difference in genetic constituents explains the great variation among the accessions. In plant breeding, this kind of information is useful for selection programmes in future include development of hybrids using distantly related plants as parents and to avoid inbreeding depression. This is the first molecular fingerprinting study of the genetic relationships of L. pumila using AFLP technique. The AFLP profiles and clusters established can be used as base for comparison of L. pumila varieties. The genetic similarity among varieties established could help future L. pumila germplasm identification, preservation and new cultivar development.
In future research, the use of a greater number of primers (Powel et al., 1996b) or some other types of molecular markers such as SSR and microsatellites may provide additional answers (Joobeur et al., 2000; Rowland et al., 2001).
Acknowledgements
The authors would like to thank the Ministry of Science, Technology and Innovation (MOSTI) for the e-Science funding support that enabled them to undertake this study. We would like to express our sincere thanks and appreciation to those who have contributed directly or indirectly to make this study a success.
References
1. Ajmone, M.P., Castiglioni, P., Fusari, F., Kuiper, M. & Motto, M. 1998. Genetic diversity and its relationship to hybrid performance in maize as revealed by RFLP and AFLP markers. Theoretical and Applied Genetics 96: 219−227.
2. Barracosa, P., Lima, M.B. & Cravador, A. 2008. Analysis of genetic diversity in Portuguese Ceratonia siliqua L.cultivars using RAPD and AFLP markers. Scientia Horticulturae 118: 189–199.
3. Bhore, S.J., Nurul, A.H. & Farida, H.S. 2009. Genetic variability based on randomly amplified polymorphic DNA in kacip fatimah (Labisia pumila Benth & Hook) collected from Melaka and Negeri Sembilan States of Malaysia. Journal of Forest Science 25: 93−100.
4. Botanga, C.J., Kling, J.G., Berner, D.K. & Timko, M.P. 2002. Genetic variability of Striga asiatica (L.) Kuntz based on AFLP analysis and host parasite interaction. Euphytica 128: 375−388.
5. Burkill, I.H. 1993. A Dictionary of the Economic Products of the Malay Peninsula v. II (I-Z). 3rd print. Kuala Lumpur: Ministry of Agriculture of Malaysia, p.1311.
6. Chen, J., Devannand, P.S., Norman, D.J., Henny, R.G. & Chao, C.C. 2004. Genetic relationships of Agloanema species and cultivars inferred AFLP markers. Annual Botanic 93(2):157−166.
7. Ehsan, K., Hawa, Z.E.J. & Sahida, A. 2011. Phenolics and flavonoids profiling and antioxidant activity of three varieties of Malaysian indigenous medicinal herb Labisia pumila Benth.Journal of Medicinal Plants Research 5(7):1200−1206.
8. Farah Fazwa, M.A., Ab. Rasip, A.G. & Lokmal, N. 2010. Screening of high total phenolic content for two varieties of Labisia pumila (Kacip Fatimah) from five different populations. Poster presented at 12th Medicinal and Aromatic Plants Seminar 2010 (APS 2010), 3–4 August 2010, Auditorium Forest Research Institute Malaysia, Kepong, Selangor.
9. Farah Fazwa, M.A., Maiden, H., Mohamad, O. & Ab. Rasip, A.G. 2012. Selection of high yielding accessions of Labisia pumila (Kacip Fatimah) for the production of superior planting stock. Journal of Tropical Medicinal Plants 13(1): 17−27.
10. Fasihuddin, A. & Hasmah, R. 1993. Kimia Hasilan Semulajadi dan Tumbuhan Ubatan. Dewan Bahasa dan Pustaka, Kuala Lumpur.
11. Gowda, B., Chandrika, K., Prasanna, K.T. & Kirana, V.C. 2010. AFLP authentication of Embelia ribes Burm. F and Embelia tsjeriam-cottama DC. International Journal of Science and Nature 1(1): 58–60.
12. Jaafar, H.Z.E., Haris, N.B.M. & Rahmat, A. 2008. Accumulation and partitioning of total phenols in two varieties of Labisia pumila Benthunder manipulation of greenhouse irradiance. Acta Horticulturae 797: 387–392.
13. Jaccard, P. 1908. Nouvelles recherché sur la distribution florale. Bull Soc Vaudoise de Sciences Naturelles44: 223–270.
14. Jamia Azdina, J., Houghton, P.J. & Miligan, S.R. 1998. Testing the Labisia pumila for aestrogenic activity using a recombinant yeast scene. Journal of Pharmaceutical Sciences 50: 79.
15. Jamia Azdina, J. 2004. Perkembangan penyelidikan dan pembangunan kacip fatimah. In Proceedings of the Seminar on Medicinal Plants and Aromatic Plants. 20–21 August 2002. (Eds): Chang, Y.S., Mastura, M. & Nurhanan, M.Y. Pp. 13–19. Forest Research Institute Malaysia (FRIM), Kepong, Malaysia.
16. Joobeur, T., Periam, N., De Vicente, M.C., King, G.J. & Arus, P. 2000. Development of a second generation linkage map for almond using RAPD and SSR markers. Genome 43: 649–655.
17. Keeratinijakal, V., Kladmook, M. & Laosatit, K. 2010. Identification and characterization of Curcuma comosa Roxb. phytoestrogens- producing plant, using AFLP markers and morphological characteristics. Journal of Medicinal Plants Research 4(24): 2651–2657.
18. Kularatne, W.W. 2000. Assessment of genetic diversity in natural oil palm (Elaeis guineensis Jacq.) populations using amplified fragment length polymorphic markers. Ph.D Theses. Universiti Kebangsaan Malaysia, Bangi.
19. Lee, S.C., Norliza, A.L., Sze, Y.L., Chew, T.L., Mohamad Roji, S. & Ramlan, A.A. 2011. Flavonoids and phenolic acids from Labisia pumila (kacip fatimah). Food Chemistry 127: 1186–1192.
20. Micheal, L.C., Brook, G.M. & Allan, E.S. 2000. Long-distance seed dispersal in plant populations. American Journal of Botany 87(9): 1217–1227.
21. Mohamad, Z. & Mustafa, A.M. 1994. Traditional Malay Medicinal Plants. Penerbit Fajar Bakti, Kuala Lumpur. 176 pp.
22. Mohd hafiz, I. & Hawa Z. E. J. 2011a. The relationship of nitrogen and C/N ratio with secondary metabolites levels and antioxidant activities in three varieties of Malaysian kacip fatimah (Labisia pumila Blume). Molecules 16: 5514–5526.
23. Mueller, U.G. & Wolfenbarger, L. 1999. AFLP genotyping and fingerprinting. Trends in Ecology and Evolution 14: 389–394.
24. Murray, M.G. & Thompson, W.F. 1980. Rapid isolation of high molecular weight DNA. Nucleic Acids Research 8: 4321–4325.
25. Nurul Arneida, H., Nur Arina, H. & Shah, F.H. 2009. Correlation between the genetic variability based on Random Amplified Polymorphic DNA (RAPD) and Random Amplified Microsatellite (RAMS) analysis and total phenolic content in garden balsam varieties (Impatiens balsamina sp.) collected from Peninsular Malaysia (TWAS). Paper Work of Regional Young Scientists Conference 2009. 2-5 November 2009, Petaling Jaya.
26. Powell, W., Machray, G.C. & Provan, J. 1996. Polymorphism revealed by simple sequence repeats. Trends in Plant Science 1: 215–222.
27. Rohlf, F.J. 1993. NTSYS-PC Numerical Taxonomy and Multivariate Analysis System, Version 1.8. Exter Publications, New York.
28. Rossi, J.A. & Singleton, V.L. 1965. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. American Journal of Enology and Viticulture16: 144-158.
29. Rowland, C.D., Saunders, S.C., Hokanson, E.L., Ogdem, A.G., Goldhirsh & Galletia, G.J. 2001. A comparison of genetic relationship measures in strawberry (Frugaria ananassa Duch.) based on ALFPS, RAPDs and pedigree data. Euphytica 117: 1–12.
30. Scalbert, A. & Williamson, G. 2000. Dietary intake and bioavailability of polyphenols. Journal of Nutrition 130: 2073–2085.
31. Sneath, P.H.A. & Sokal, R.R. 1973. Numerical Taxomony. Freeman, San Francisco.
32. Velioglu, Y.S., Mazza, G., Gao, L. & Oomah, B.D. 1998. Antioxidant activity and total phenolics in selected fruits, vegetables and grain products. Journal of Agricultural and Food Chemistry 46: 4113–4117.
33. Vimala, S. 2010. Total phenolic content in herbal plants. [Pers. Comm, 10 September 2010.
34. Vos, P., Hogers, R., Bleeker, M., Reijans, M., Van De Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M. & Zabeau, M. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407−4414.
35. Wickneswari, R., Lee, C.T, Norwati, M. & Boyle, T.J.B. 2000. Impact of logging on genetic diversity in humid tropical forests. In Matyas, C. (Ed.). Forest Genetics and Sustainability pp.171–181. Kluwer Academic Publishers, The Netherlands.
36. Wong, S.P., Leong, L.P. & Koh, J.H.W. 2006. Antioxidant activities of aqueous extracts of selected plants. Food Chemistry 99: 775−783.
37. Yang, J., Qian, Z., Liu, Z., Li, S., Sun, G. & Zhao, G. 2007. Genetic diversity and geographical differentiation of Dipteronia oliv. (Aceraceae) endemic to China as revealed by AFLP analysis. Biochemical Systematics and Ecology 35: 593−599.
Cite this paper
APA
Md Ariff, F. F., Hashim, S. S., Haja, M., & Osman, M. (2013). An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers. Open Science Repository Agriculture, Online(open-access), e70081945. doi:10.7392/Agriculture.70081945
MLA
Md Ariff, Farah Fazwa et al. “An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers.” Open Science Repository Agriculture Online.open-access (2013): e70081945.
Chicago
Md Ariff, Farah Fazwa, Siti Salwana Hashim, Maideen Haja, and Mohamad Osman. “An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers.” Open Science Repository Agriculture Online, no. open-access (March 22, 2013): e70081945. http://www.open-science-repository.com/an-assessment-of-genetic-relationship-among-superior-accessions-of-labisia-pumila.html.
Harvard
Md Ariff, F.F. et al., 2013. An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers. Open Science Repository Agriculture, Online(open-access), p.e70081945. Available at: http://www.open-science-repository.com/an-assessment-of-genetic-relationship-among-superior-accessions-of-labisia-pumila.html.
Science
1. F. F. Md Ariff, S. S. Hashim, M. Haja, M. Osman, An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers, Open Science Repository Agriculture Online, e70081945 (2013).
Nature
1. Md Ariff, F. F., Hashim, S. S., Haja, M. & Osman, M. An Assessment of Genetic Relationship Among Superior Accessions of Labisia Pumila Analized by Amplified Fragment Length Polymorphism (AFLP) Markers. Open Science Repository Agriculture Online, e70081945 (2013).
doi
Research registered in the DOI resolution system as: 10.7392/Agriculture.70081945.

This work is licensed under a Creative Commons Attribution 3.0 Unported License.